Code
import os
if "KERAS_BACKEND" not in os.environ:
# set this to "torch", "tensorflow", or "jax"
os.environ["KERAS_BACKEND"] = "jax"
import matplotlib.pyplot as plt
import numpy as np
import bayesflow as bf
import jsonimport os
if "KERAS_BACKEND" not in os.environ:
# set this to "torch", "tensorflow", or "jax"
os.environ["KERAS_BACKEND"] = "jax"
import matplotlib.pyplot as plt
import numpy as np
import bayesflow as bf
import json\[\begin{equation} \begin{aligned} \gamma & \sim \text{Uniform}(0.5, 1) \\ y_{iq} & \sim \begin{cases} \text{Bernoulli}(\gamma) & \text{if } t_q = a \\ \text{Bernoulli}(1-\gamma) & \text{if } t_q = b \\ \text{Bernoulli}(0.5) & \text{otherwise}, \\ \end{cases} \end{aligned} \end{equation}\] where \(t_q\) is determined by the take the best decision rule defined below.
with open(os.path.join("data", "StopSearchData.json")) as f:
data = json.load(f)
data.keys()dict_keys(['m', 'nc', 'nq', 'ns', 'p', 'v', 'x', 'y'])First, we define the “take the best” (TTB) decision rule: For each question, we loop over its cues in order of their validity (the questions are already ordered in the data). As soon as a cue prefers decision ‘a’ or ‘b’, the decision is determined.
def ttb(nq: int, nc: int, p: np.ndarray, m: np.ndarray):
output = np.zeros(nq) # 0: guessing
for q in range(nq):
for c in range(nc):
cue_a = m[p[q][0]-1][c]
cue_b = m[p[q][1]-1][c]
if cue_a > cue_b:
output[q] = 1 # decision a
break
elif cue_a < cue_b:
output[q] = 2 # decision b
break
return output.astype(int)Applying the decision rule to the data produces the “decision a” for each question according to TTB: This is because the recorded data had been already recoded such that choice “a” always corresponds to the choice according to the TTB rule.
decisions = ttb(data['nq'], data['nc'], data['p'], data['m'])
decisionsarray([1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1])Next, we can define the prior and a likelihood. The only source of alleatoric uncertainty in our simulator comes from the parameter \(\gamma\), which indicates the probability of responding “a” if the TTB decision rule indicates to respond “a”, and vice versa.
The shape of the resulting data is (number of participants, number of questions).
def prior():
return dict(gamma = np.random.uniform(low=0.5, high=1))
def likelihood(
gamma: float,
ns: int = data['ns'],
nq: int = data['nq'],
ttb = decisions
):
probs = np.array([0.5, gamma, 1-gamma])
probs = probs[ttb][np.newaxis]
probs = np.tile(probs, reps=[ns, 1])
y = np.random.binomial(n=1, p=probs, size=(ns, nq))
return dict(y=y)
simulator = bf.make_simulator([prior, likelihood])We will approximate the posterior of \(\gamma\), given the observed decisions \(y\).
adapter = (bf.Adapter()
.constrain("gamma", lower=0.5, upper=1)
.rename("gamma", "inference_variables")
.rename("y", "summary_variables")
)workflow = bf.BasicWorkflow(
simulator=simulator,
adapter=adapter,
inference_network=bf.networks.CouplingFlow(),
summary_network=bf.networks.DeepSet(),
initial_learning_rate=1e-3
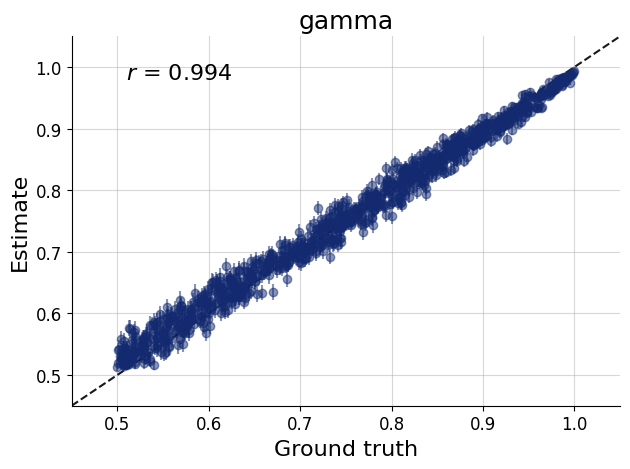
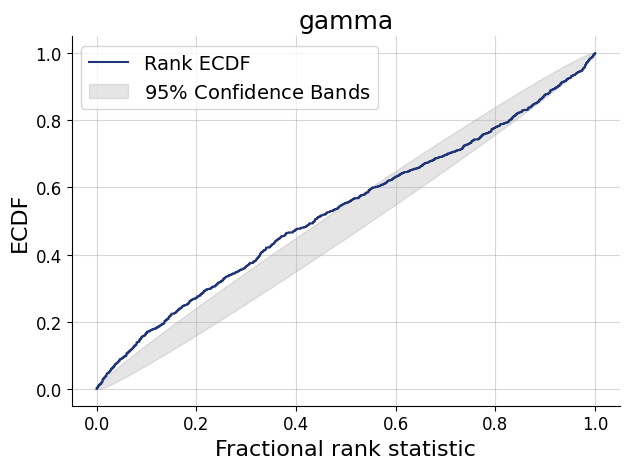
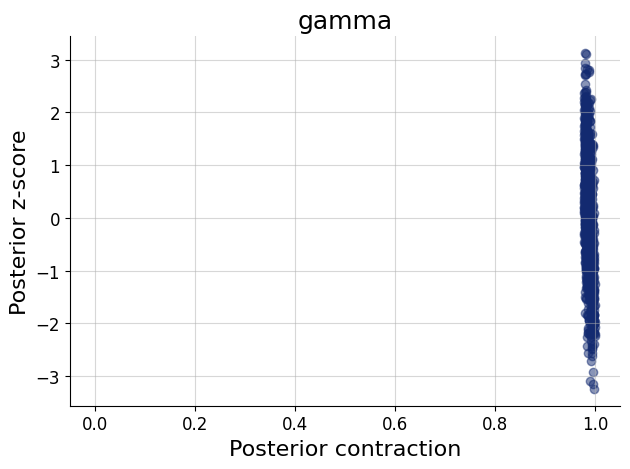
)history = workflow.fit_online(epochs=50, num_batches_per_epoch=500, batch_size=512)test_data = simulator.sample(1000)figs=workflow.plot_default_diagnostics(test_data=test_data, num_samples=500)
Now we can apply to the model to real data (Lee & Wagenmakers, 2013).
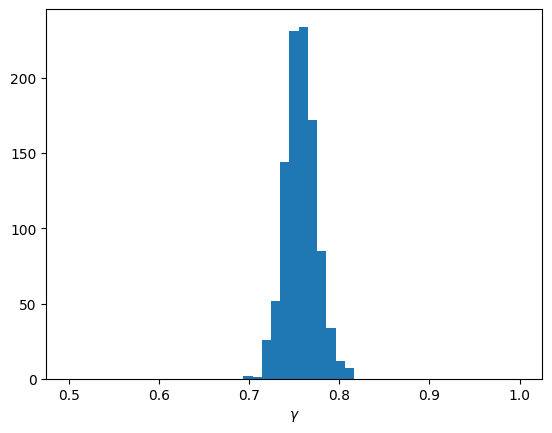
inference_data = dict(y = np.array(data['y'])[np.newaxis])samples = workflow.sample(num_samples=1000, conditions=inference_data)f=plt.hist(samples['gamma'].flatten(), bins=np.linspace(0.5, 1, 50))
f=plt.xlabel(r"$\gamma$")