Code
import os
if "KERAS_BACKEND" not in os.environ:
# set this to "torch", "tensorflow", or "jax"
os.environ["KERAS_BACKEND"] = "jax"
import matplotlib.pyplot as plt
import numpy as np
import keras
import bayesflow as bfimport os
if "KERAS_BACKEND" not in os.environ:
# set this to "torch", "tensorflow", or "jax"
os.environ["KERAS_BACKEND"] = "jax"
import matplotlib.pyplot as plt
import numpy as np
import keras
import bayesflow as bf\[\begin{equation} \begin{aligned} c & \sim \text{Beta}(1, 1) \\ r & \sim \text{Beta}(1, 1) \\ u & \sim \text{Beta}(1, 1) \\ \theta_1 & \leftarrow cr \\ \theta_2 & \leftarrow (1-c)u^2 \\ \theta_3 & \leftarrow 2u(1-c)(1-u)\\ \theta_4 & \leftarrow (1-c)(1-u)^2 \\ k & \sim \text{Multinomial}(\theta, n) \end{aligned} \end{equation}\]
def context():
return dict(n=np.random.randint(10, 1001))
def prior():
return dict(
c = np.random.beta(a=1, b=1),
r = np.random.beta(a=1, b=1),
u = np.random.beta(a=1, b=1)
)
def likelihood(c, r, u, n):
theta = [c*r, (1-c)*u**2, 2*u*(1-c)*(1-u), c*(1-r) + (1-c)*(1-u)**2]
k = np.random.multinomial(n=n, pvals=theta)
return dict(k=k)
simulator = bf.make_simulator([context, prior, likelihood])(10, 4)We will use the BasicWorkflow to aproximate the posterior of \(c\), \(r\), and \(u\), given sample size \(n\) and observed responses \(k\).
We will also make sure that the constraints of the parameters (which all live on the interval between 0 and 1) are respected.
adapter = (bf.Adapter()
.constrain(["c", "r", "u"], lower=0, upper=1)
.concatenate(["c", "r", "u"], into="inference_variables")
.concatenate(["n", "k"], into="inference_conditions")
)
workflow = bf.BasicWorkflow(
simulator=simulator,
adapter=adapter,
inference_network=bf.networks.CouplingFlow()
)history=workflow.fit_online(epochs=20, num_batches_per_epoch=100, batch_size=256)test_data = simulator.sample(1000)
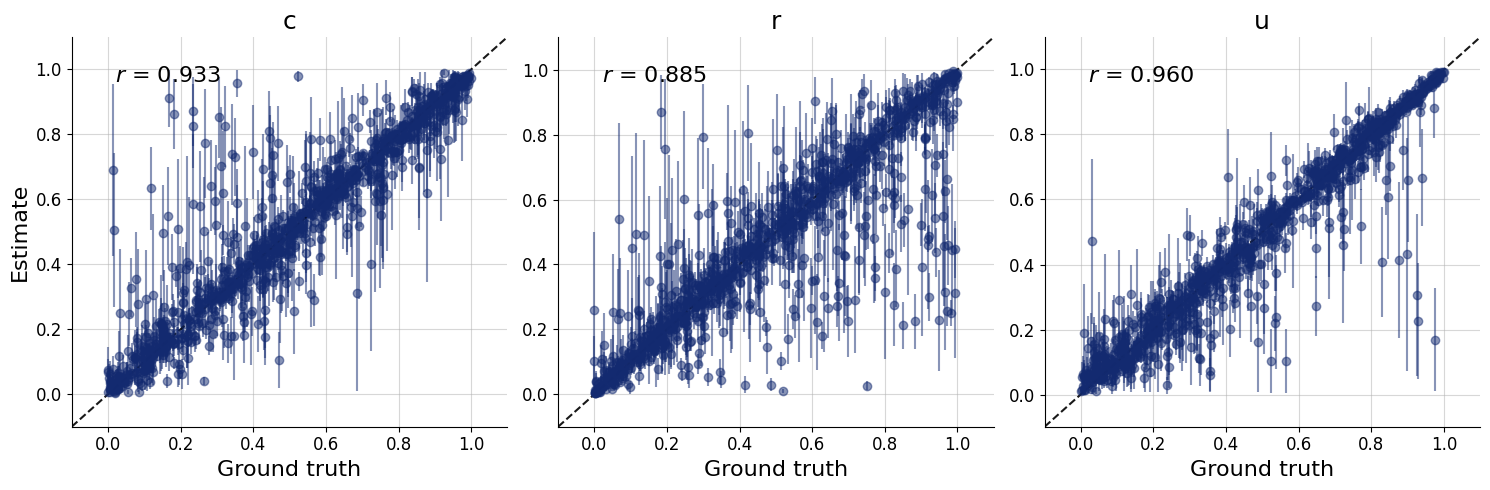
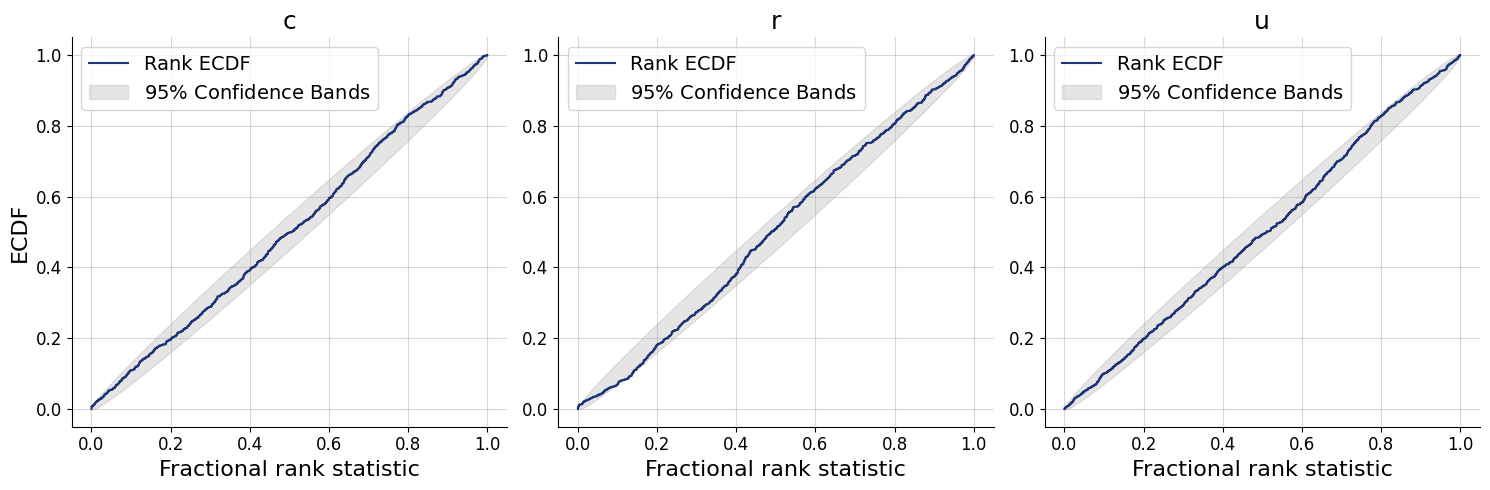
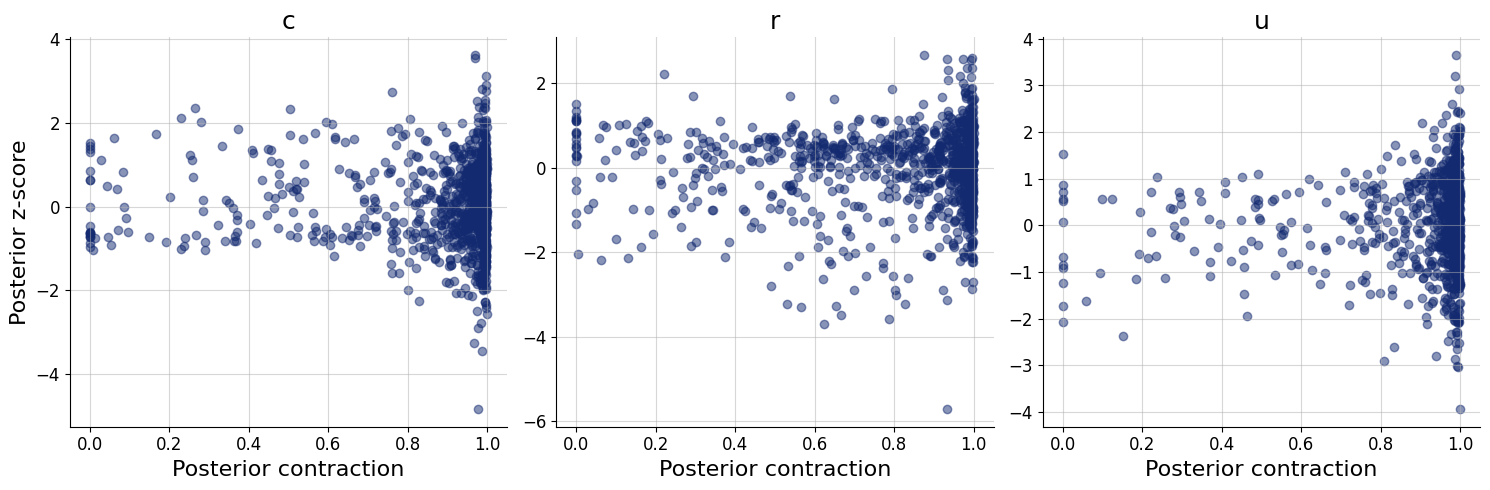
figs=workflow.plot_default_diagnostics(test_data=test_data, num_samples=500)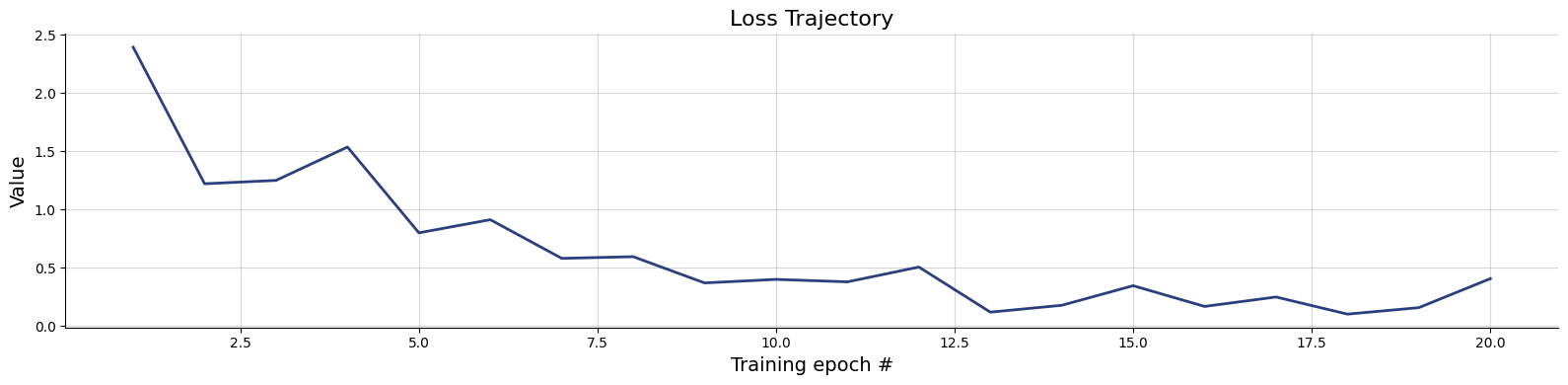
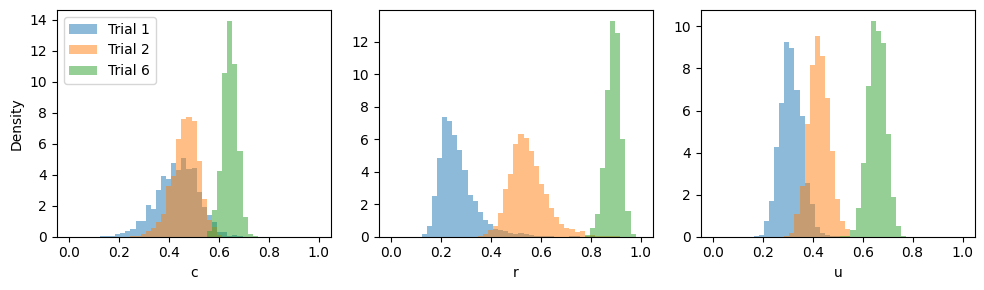
Here we will obtain the posterior distributions for trials 1, 2, and 6 from Riefer et al. (2002) as reported by Lee & Wagenmakers (2013).
k = np.array([
[45, 24, 97, 254],
[106, 41, 107, 166],
[243, 64, 65, 48]
])
inference_data = dict(k=k, n=np.sum(k, axis=-1)[..., np.newaxis])posterior_samples = workflow.sample(num_samples=2000, conditions=inference_data)plt.rcParams['figure.figsize'] = [10, 3]
fig, axes = plt.subplots(ncols=3)
axes = axes.flatten()
labels = ["Trial 1", "Trial 2", "Trial 6"]
for dat in range(len(labels)):
for i, (par, samples) in enumerate(posterior_samples.items()):
axes[i].hist(samples[dat].flatten(), density=True, alpha=0.5, bins = np.linspace(0, 1, 50))
axes[i].set_xlabel(par)
axes[0].legend(labels)
axes[0].set_ylabel("Density")
fig.tight_layout()