Code
import os
if "KERAS_BACKEND" not in os.environ:
# set this to "torch", "tensorflow", or "jax"
os.environ["KERAS_BACKEND"] = "jax"
import matplotlib.pyplot as plt
import numpy as np
import bayesflow as bf
import jsonimport os
if "KERAS_BACKEND" not in os.environ:
# set this to "torch", "tensorflow", or "jax"
os.environ["KERAS_BACKEND"] = "jax"
import matplotlib.pyplot as plt
import numpy as np
import bayesflow as bf
import jsonHere we will allow for individual differences between participants. The simplest way to start is to estimate the parameter values for each participant independently. Hence we estimate two parameters per participant. It may seem that we have to extend the model. However, since we assume that the participants are independent, we may as well simplify the model to assume data from a single participant only, and apply it to each participant in independently.
We use the category learning data from the “Condensation B” condition from Kruschke (1993) as reported by Lee & Wagenmakers (2013).
with open(os.path.join("data", "KruschkeData.json")) as f:
data = json.load(f)
data.keys()dict_keys(['a', 'd1', 'd2', 'n', 'nstim', 'nsubj', 'y'])We will reuse the prior and simulator from the first example of the GCM. The only difference is that we will remove the nsubj argument, and in so doing simplifying the simulation so that every data set contains data from a single participant.
def prior():
return dict(
c = np.random.exponential(scale=1),
w = np.random.uniform(low=0, high=1)
)
def likelihood(c, w, n=data['n'], nstim=data['nstim'], d1=np.array(data['d1']), d2=np.array(data['d2']), a=np.array(data['a'])):
b = 0.5
s = np.exp( - c * (w * d1 + (1-w) * d2))
r = np.zeros(nstim)
for i in range(nstim):
isA = a == 1
isB = a != 1
numerator = b * np.sum(s[i,isA])
denominator = numerator + (1-b) * np.sum(s[i,isB])
r[i] = numerator / denominator
y = np.random.binomial(n=n, p=r, size=nstim)
return dict(y=y)
simulator = bf.make_simulator([prior, likelihood])We will use BasicWorkflow again. The task is to predict the posterior distribution of the parameters \(c\) and \(w\), given the observed data \(y\). Since \(y\) is of fixed size, we might not need a summary network.
adapter = (bf.Adapter()
.constrain("c", lower=0)
.constrain("w", lower=0, upper=1)
.concatenate(["c", "w"], into="inference_variables")
.rename("y", "inference_conditions")
)workflow = bf.BasicWorkflow(
simulator=simulator,
adapter=adapter,
inference_network=bf.networks.CouplingFlow(),
)history=workflow.fit_online(epochs=50, batch_size=256)test_data = simulator.sample(1000)figs = workflow.plot_default_diagnostics(test_data=test_data, num_samples=500)
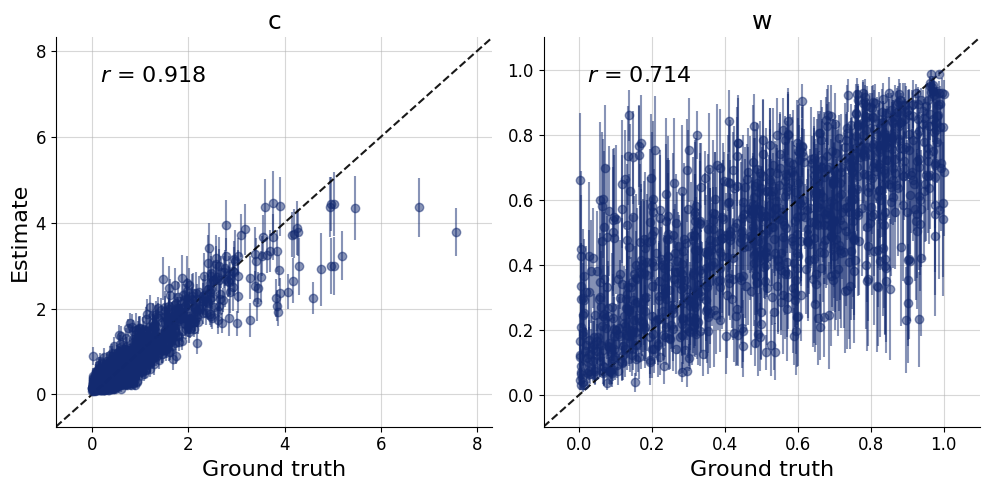
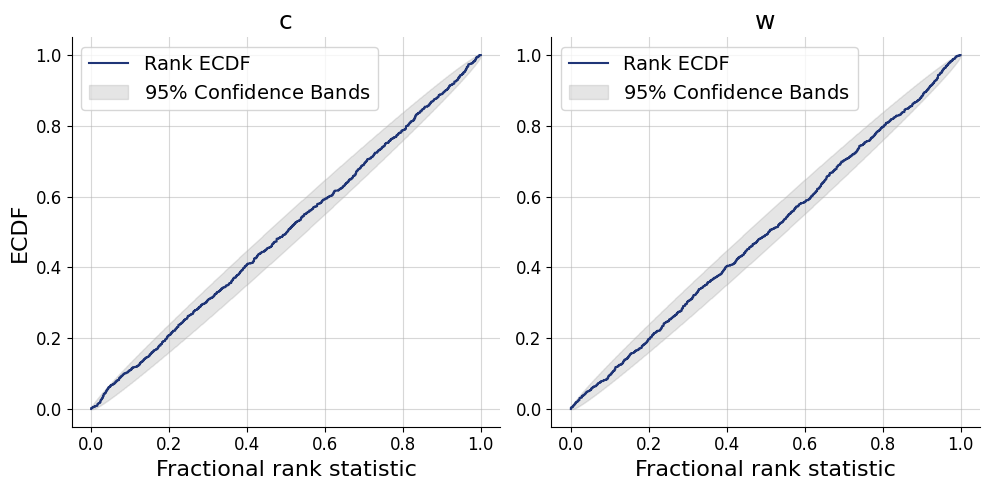
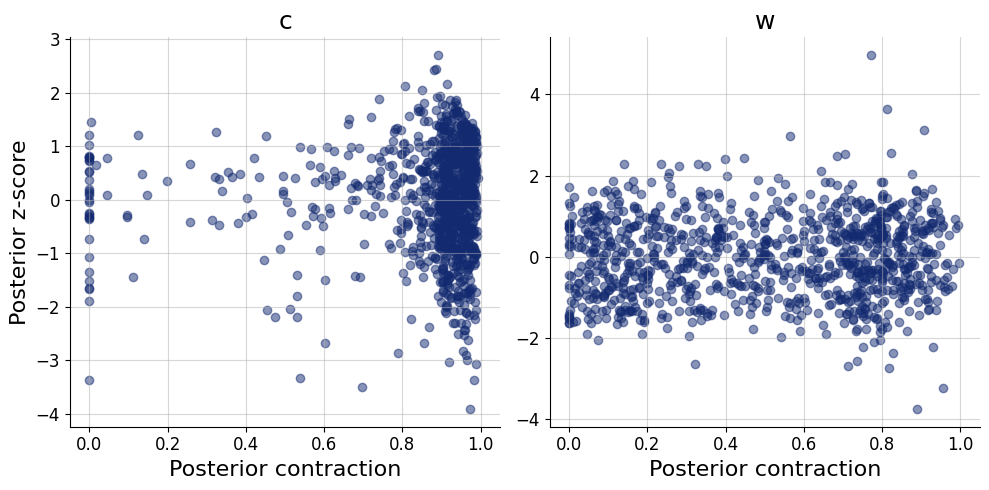
Now, we can use the approximator to obtain our posterior distributions of \(c\) and \(w\) for all 40 participants in the data set.
inference_data = dict(y = np.array(data['y']).transpose())
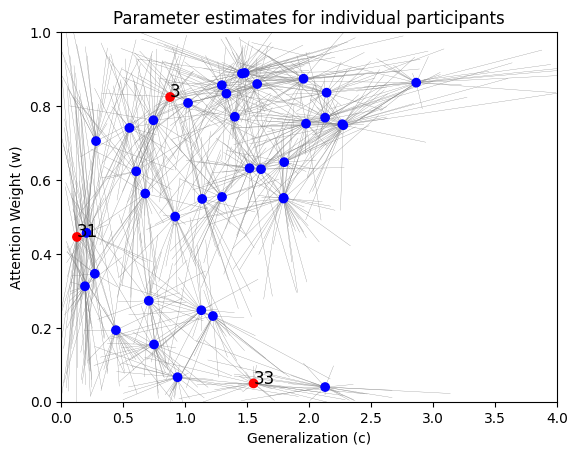
inference_data['y'].shape(40, 8)posterior_samples = workflow.sample(num_samples=2000, conditions=inference_data)posterior_means = {par: np.mean(samples, axis=1) for par, samples in posterior_samples.items()}highlight = [2, 30, 32]
cols = ["blue" if i not in highlight else "red" for i in range(data['nsubj'])]
plt.scatter(
posterior_means['c'],
posterior_means['w'],
color=cols, zorder=2)
for p in range(data['nsubj']):
if p in highlight:
plt.text(
posterior_means['c'][p],
posterior_means['w'][p],
str(p+1), zorder=3, fontsize=12)
for l in range(25):
x = [posterior_means['c'][p],
posterior_samples['c'][p,l]
]
y = [posterior_means['w'][p],
posterior_samples['w'][p,l]
]
plt.plot(x, y, color="gray", linewidth=0.2, zorder=0)
plt.xlim(0, 4)
plt.xlabel("Generalization (c)")
plt.ylim(0, 1)
plt.ylabel("Attention Weight (w)")
f=plt.title("Parameter estimates for individual participants")