Code
import os
if "KERAS_BACKEND" not in os.environ:
# set this to "torch", "tensorflow", or "jax"
os.environ["KERAS_BACKEND"] = "jax"
import matplotlib.pyplot as plt
import numpy as np
import bayesflow as bf
import jsonimport os
if "KERAS_BACKEND" not in os.environ:
# set this to "torch", "tensorflow", or "jax"
os.environ["KERAS_BACKEND"] = "jax"
import matplotlib.pyplot as plt
import numpy as np
import bayesflow as bf
import jsonThe basic model has three parameters - generalization (c), attention weight (w), and bias (b). In this example, we assume no bias between the categories, so b will be fixed to \(\frac{1}{2}\).
\[\begin{equation} \begin{aligned} c & \sim \text{Exponential}(0.5) \\ w & \sim \text{Uniform}(0, 1) \\ b & \leftarrow \frac{1}{2} \\ s_{ij} & \leftarrow \exp\left[- c \left(w d_{ij}^{(1)} + (1-w) d_{ij}^{(2)}\right)\right] \\ r_i & \leftarrow \frac{b \sum_j a_j s_{ij}}{b \sum_j a_j s_{ij} + (1-b) \sum_j (1-a_j) s_{ij}} \\ y_i & \sim \text{Binomial}(r_i, t) \end{aligned} \end{equation}\]
We use the category learning data from the “Condensation B” condition reported by Kruschke (1993).
with open(os.path.join("data", "KruschkeData.json")) as f:
data = json.load(f)
data.keys()dict_keys(['a', 'd1', 'd2', 'n', 'nstim', 'nsubj', 'y'])The element y contains the array of responses \(y_{ik}\) for the trial \(i \in \{1, \dots, 8\}\) and participant \(k \in \{1, \dots, 40\}\).
np.array(data['y']).shape(8, 40)d1 and d2 contain the physical distances between the 8 stimuli on the along the two dimensions, such that
\[\begin{equation} d_{ij}^{(k)} = | p_{ik} - p_{jk} | \end{equation}\]
The indicator vector a contains information whether the stimulus \(a_i\) is an element from Category A or an element from Category B.
Here we will assume that the design of the experiment (i.e., number of trials, number of stimuli, number of participants, nature of the stimuli) is fixed. That is, we will only amortize over the observation outcomes.
def prior():
return dict(
c = np.random.exponential(scale=1),
w = np.random.uniform(low=0, high=1)
)
def likelihood(c, w, n=data['n'], nstim=data['nstim'], nsubj=data['nsubj'], d1=np.array(data['d1']), d2=np.array(data['d2']), a=np.array(data['a'])):
b = 0.5
s = np.exp( - c * (w * d1 + (1-w) * d2))
r = np.zeros((nsubj, nstim))
for i in range(nstim):
isA = a == 1
isB = a != 1
numerator = b * np.sum(s[i,isA])
denominator = numerator + (1-b) * np.sum(s[i,isB])
r[:, i] = numerator / denominator
y = np.random.binomial(n=n, p=r, size=(nsubj, nstim))
return dict(y=y)
simulator = bf.make_simulator([prior, likelihood])adapter = (bf.Adapter()
.constrain("c", lower=0)
.constrain("w", lower=0, upper=1)
.concatenate(["c", "w"], into="inference_variables")
.rename("y", "summary_variables")
)workflow=bf.BasicWorkflow(
simulator=simulator,
adapter=adapter,
inference_network=bf.networks.CouplingFlow(),
summary_network=bf.networks.DeepSet(),
initial_learning_rate=1e-3
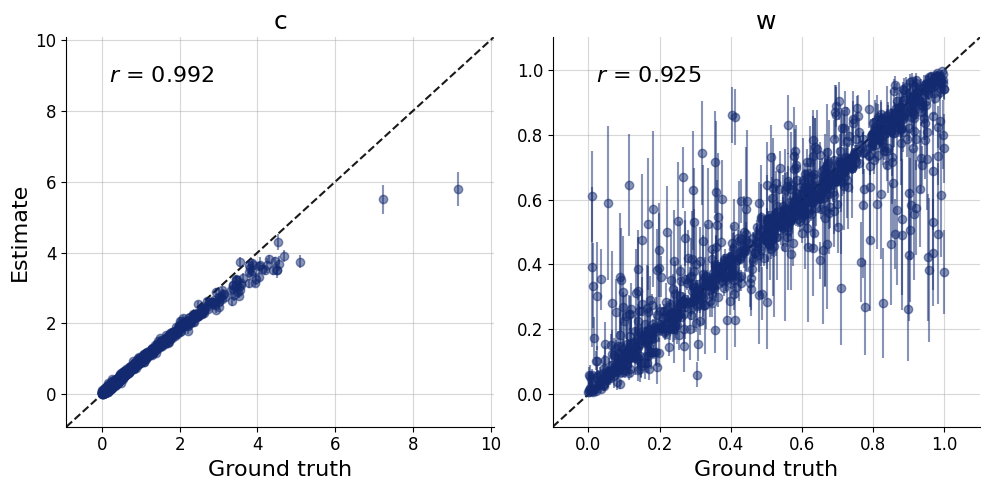
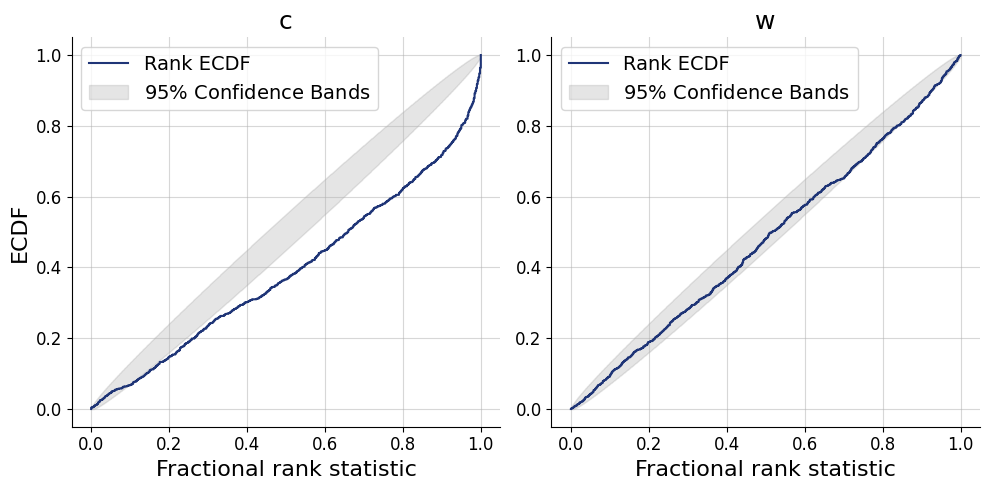
)history=workflow.fit_online(batch_size=256)test_data = simulator.sample(1000)figs = workflow.plot_default_diagnostics(test_data=test_data, num_samples=500)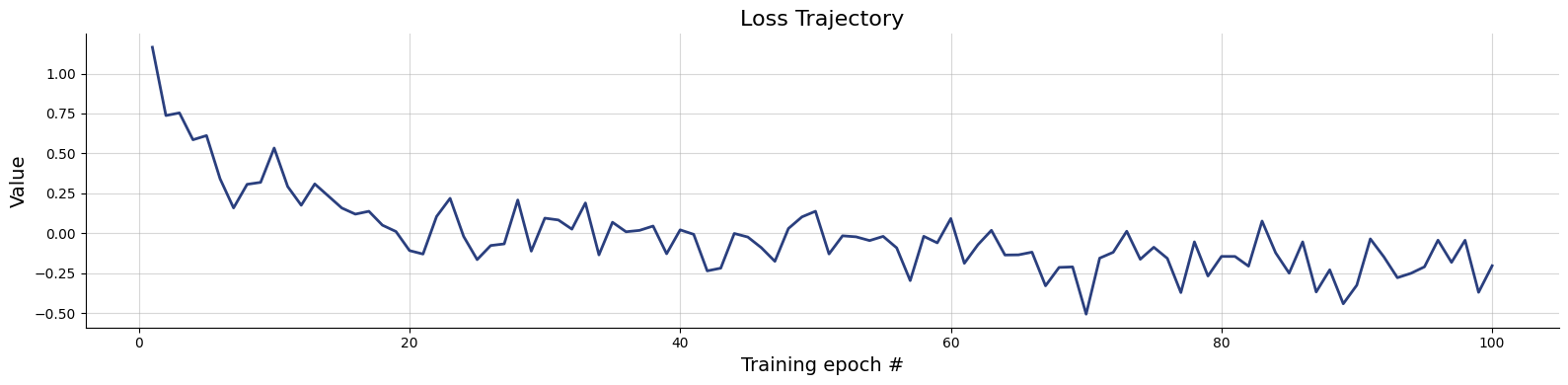
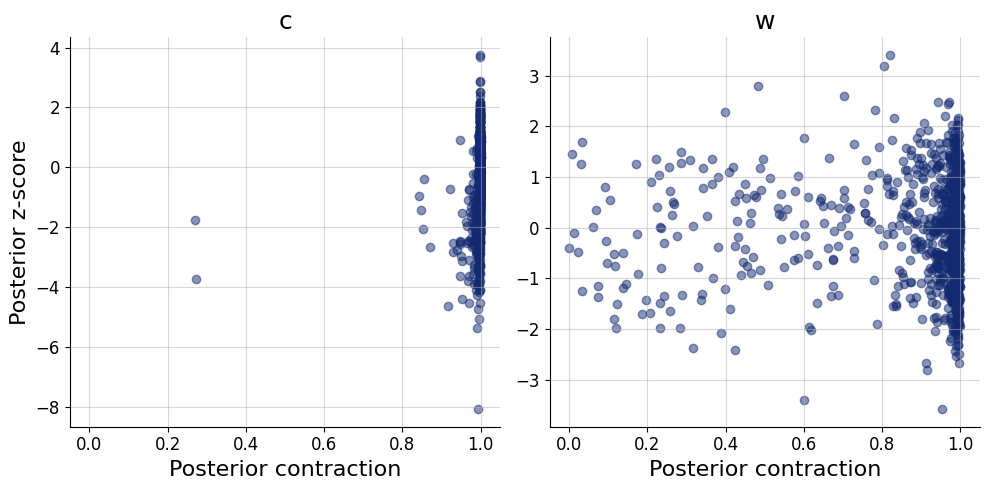
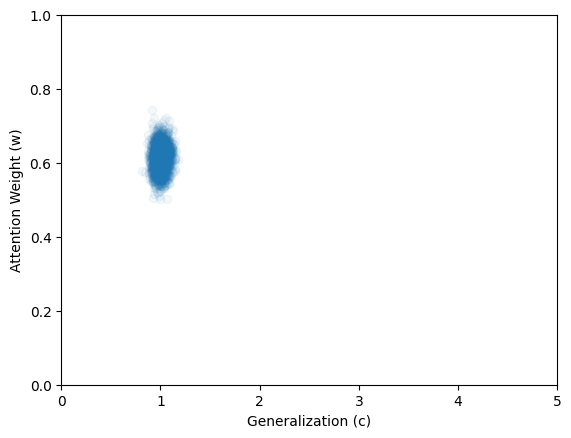
Here we apply the networks to the data from Kruschke (1993) as reported by Lee & Wagenmakers (2013). The quantity of interest is the joint posterior distribution of parameters \(c\) and \(w\).
inference_data = dict(y = np.array(data['y']).transpose()[np.newaxis,...])posterior_samples = workflow.sample(num_samples=2000, conditions=inference_data)plt.scatter(
x = posterior_samples['c'],
y = posterior_samples['w'],
alpha=0.05
)
plt.xlim([0, 5])
plt.ylim([0, 1])
plt.xlabel("Generalization (c)")
f=plt.ylabel("Attention Weight (w)")