Code
import osif "KERAS_BACKEND" not in os.environ:# set this to "torch", "tensorflow", or "jax" "KERAS_BACKEND" ] = "jax" import matplotlib.pyplot as pltimport numpy as npimport bayesflow as bf
INFO:bayesflow:Using backend 'jax'
\[\begin{equation}
\begin{aligned}
\alpha & \sim \text{Beta}(1, 1) \\
\beta & \sim \text{Beta}(1, 1) \\
\theta_i' & \leftarrow \exp(-\alpha t_i) + \beta \\
\theta_i & \leftarrow \begin{cases}
0 & \text{if } \theta_i' < 0 \\
1 & \text{if } \theta_i' > 1 \\
\theta_i' & \text{otherwise}
\end{cases} \\
k_i & \sim \text{Binomial}(\theta_i, 18)
\end{aligned}
\end{equation}\]
Simulator
Code
def prior():= np.random.beta(a= 1 , b= 1 )= np.random.beta(a= 1 , b= 1 )return dict (alpha= alpha, beta= beta)def likelihood(alpha, beta, t= np.array([1 , 2 , 4 , 7 , 12 , 21 , 35 , 59 , 99 ])):= np.exp(- alpha* t) + beta= np.clip(theta, a_min= 0 , a_max= 1 )= np.random.binomial(n= 18 , p= theta, size= (3 , len (theta)))return dict (k= k)= bf.make_simulator([prior, likelihood])
Approximator
Code
= ("alpha" , "beta" ], lower= 0 , upper= 1 )"alpha" , "beta" ], into= "inference_variables" )"k" , "summary_variables" )
Code
= bf.BasicWorkflow(= simulator,= adapter,= bf.networks.CouplingFlow(),= bf.networks.DeepSet(),= 1e-3
Training
Code
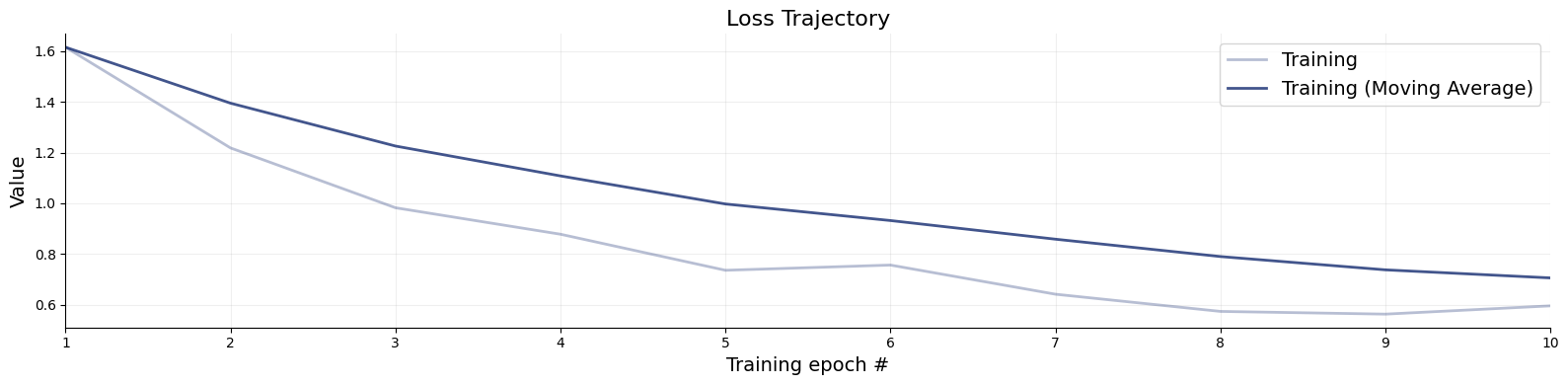
= workflow.fit_online(epochs= 10 , num_batches_per_epoch= 100 , batch_size= 1024 )
Code
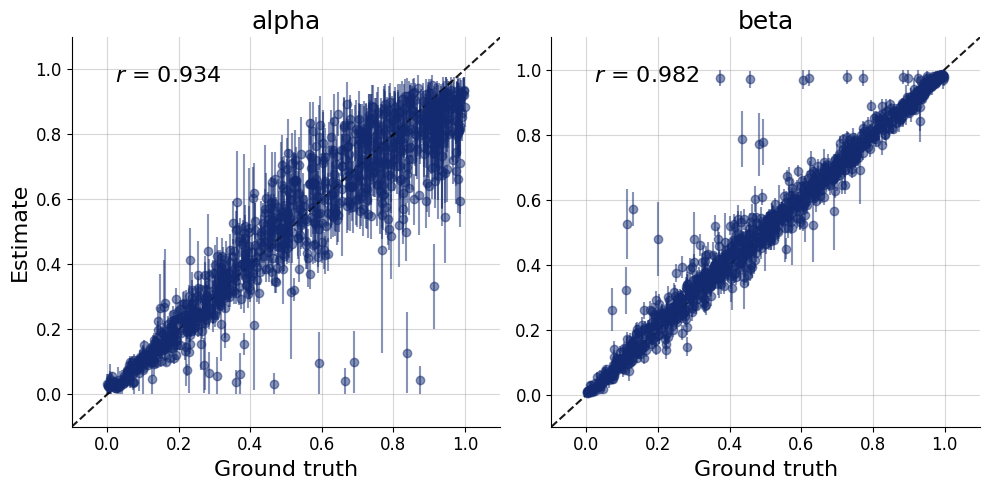
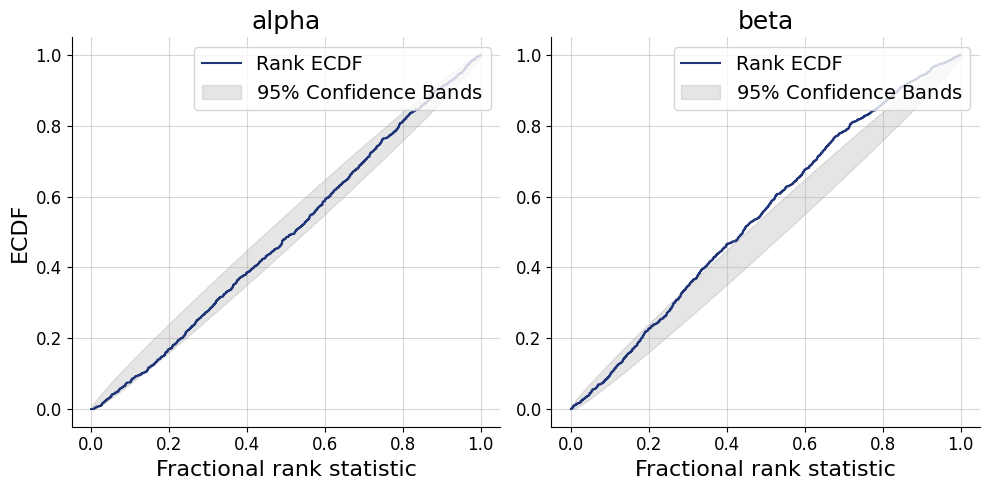
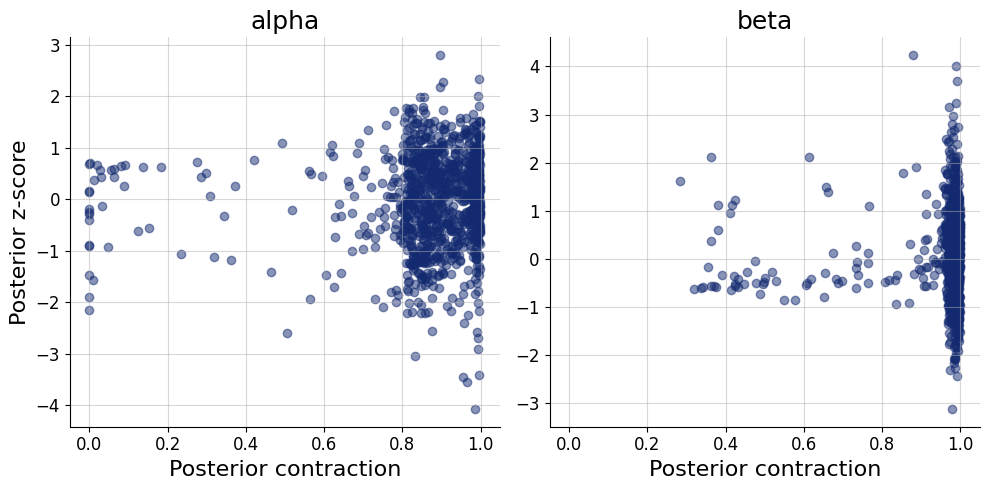
= simulator.sample(1000 )= workflow.plot_default_diagnostics(test_data= test_data, num_samples= 500 )
Inference
We use the data from three participants by Rubin et al. (1999 ) as reported by Lee & Wagenmakers (2013 ) .
Code
= [[18 , 18 , 16 , 13 , 9 , 6 , 4 , 4 , 4 ],17 , 13 , 9 , 6 , 4 , 4 , 4 , 4 , 4 ],14 , 10 , 6 , 4 , 4 , 4 , 4 , 4 , 4 ]]= dict (k= np.array(k)[np.newaxis])
Code
= workflow.sample(num_samples= 2000 , conditions= inference_data)
Code
= samples["alpha" ], = samples["beta" ], = 0.3 )0 , 1 ])0 , 1 ])"Decay Rate $ \\ alpha$" )"Baseline $ \\ beta$" )None
References
Lee, M. D., & Wagenmakers, E.-J. (2013). Bayesian Cognitive Modeling : A Practical Course . Cambridge University Press.
Rubin, D. C., Hinton, S., & Wenzel, A. (1999). The precise time course of retention. Journal of Experimental Psychology: Learning, Memory, and Cognition , 25 (5), 1161.