Code
import os
if "KERAS_BACKEND" not in os.environ:
# set this to "torch", "tensorflow", or "jax"
os.environ["KERAS_BACKEND"] = "jax"
import matplotlib.pyplot as plt
import numpy as np
import bayesflow as bf
import kerasimport os
if "KERAS_BACKEND" not in os.environ:
# set this to "torch", "tensorflow", or "jax"
os.environ["KERAS_BACKEND"] = "jax"
import matplotlib.pyplot as plt
import numpy as np
import bayesflow as bf
import kerasHere we will compare two models. The null model assumes that proportion is the same for two binomial data sets
\[\begin{equation} \begin{aligned} s_1 & \sim \text{Binomial}(\theta_1, n_1) \\ s_2 & \sim \text{Binomial}(\theta_1, n_2) \\ \theta_1 & \sim \text{Beta}(1, 1), \end{aligned} \end{equation}\] whereas the alternative model assumes that the two groups are associated with different binomial rates:
\[\begin{equation} \begin{aligned} s_1 & \sim \text{Binomial}(\theta_1, n_1) \\ s_2 & \sim \text{Binomial}(\theta_1, n_2) \\ \theta_1 & \sim \text{Beta}(1, 1) \\ \theta_2 & \sim \text{Beta}(1, 1). \end{aligned} \end{equation}\]
We will also amortize over different sample sizes for each group.
def context():
n = np.random.randint(1000, 10_000, size=2)
return dict(n = n)
def prior_null():
theta = np.random.beta(a=1, b=1)
return dict(theta = np.array([theta, theta]))
def prior_alternative():
theta = np.random.beta(a=1, b=1, size=2)
return dict(theta = theta)
def likelihood(theta, n):
s = np.random.binomial(n=n, p=theta)
return dict(s=s)
simulator_null = bf.make_simulator([context, prior_null, likelihood])
simulator_alternative = bf.make_simulator([context, prior_alternative, likelihood])
simulator = bf.simulators.ModelComparisonSimulator(
simulators=[simulator_null, simulator_alternative],
use_mixed_batches=True)adapter = (
bf.Adapter()
.concatenate(['n', 's'], into="classifier_conditions")
.drop('theta')
)classifier_network = keras.Sequential([
keras.layers.Dense(32, activation="gelu")
for _ in range(8)
])
approximator = bf.approximators.ModelComparisonApproximator(
num_models=2,
classifier_network=classifier_network,
adapter=adapter)epochs = 30
num_batches = 100
batch_size = 512
learning_rate = keras.optimizers.schedules.CosineDecay(5e-4, decay_steps=epochs*num_batches)
optimizer = keras.optimizers.Adam(learning_rate=learning_rate, clipnorm=1.0)
approximator.compile(optimizer=optimizer)history = approximator.fit(
epochs=epochs,
num_batches=num_batches,
batch_size=batch_size,
simulator=simulator,
adapter=adapter
)f=bf.diagnostics.plots.loss(history=history)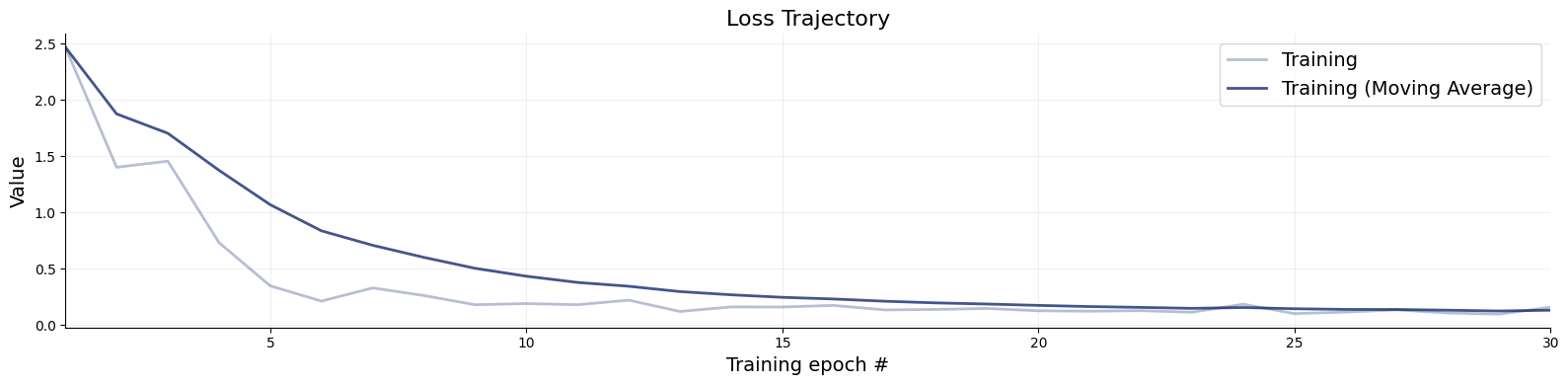
test_data=simulator.sample(5_000)true_models=test_data["model_indices"]
pred_models=approximator.predict(conditions=test_data)f=bf.diagnostics.plots.mc_calibration(
pred_models=pred_models,
true_models=true_models,
model_names=[r"$\mathcal{M}_0$",r"$\mathcal{M}_1$"],
)INFO:matplotlib.mathtext:Substituting symbol M from STIXNonUnicode
INFO:matplotlib.mathtext:Substituting symbol M from STIXNonUnicode
INFO:matplotlib.mathtext:Substituting symbol M from STIXNonUnicode
INFO:matplotlib.mathtext:Substituting symbol M from STIXNonUnicode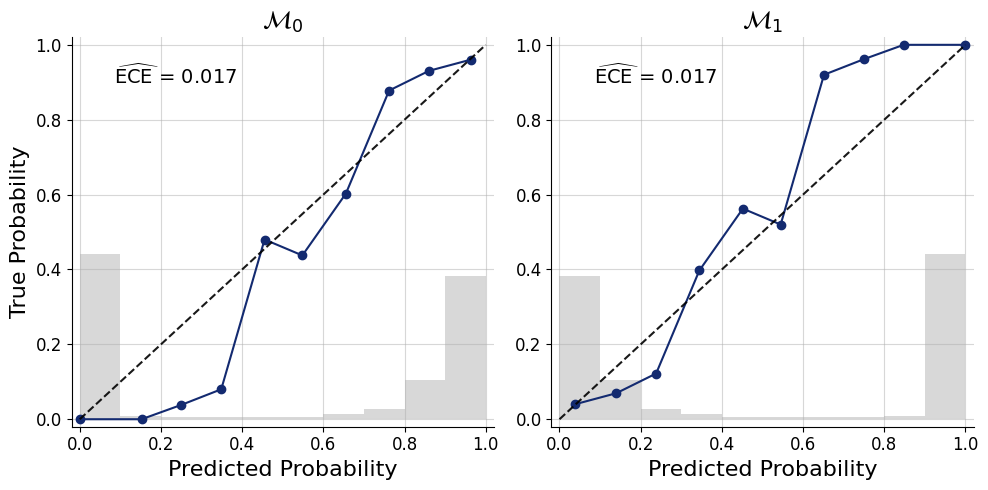
f=bf.diagnostics.plots.mc_confusion_matrix(
pred_models=pred_models,
true_models=true_models,
model_names=[r"$\mathcal{M}_0$",r"$\mathcal{M}_1$"],
normalize="true"
)INFO:matplotlib.mathtext:Substituting symbol M from STIXNonUnicode
INFO:matplotlib.mathtext:Substituting symbol M from STIXNonUnicode
INFO:matplotlib.mathtext:Substituting symbol M from STIXNonUnicode
INFO:matplotlib.mathtext:Substituting symbol M from STIXNonUnicode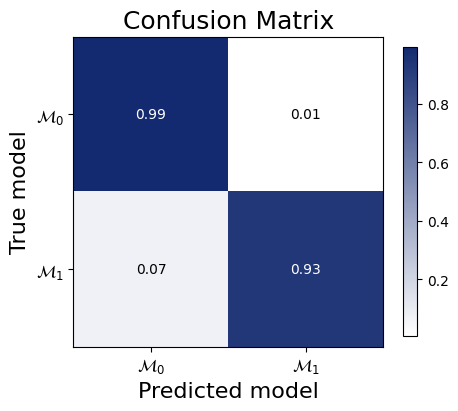
inference_data = dict(n = np.array([[5416, 9072]]), s = np.array([[424, 777]]))pred_models = approximator.predict(conditions=inference_data)[0]
pred_modelsarray([0.9855641 , 0.01443596], dtype=float32)pred_models[1]/pred_models[0]0.014647411