Code
import osif "KERAS_BACKEND" not in os.environ:# set this to "torch", "tensorflow", or "jax" "KERAS_BACKEND" ] = "jax" import matplotlib.pyplot as pltimport numpy as npimport bayesflow as bf
INFO:bayesflow:Using backend 'jax'
\[\begin{equation}
\begin{aligned}
\gamma & \sim \text{Exponential}(1) \\
\beta & \sim \text{Exponential}(1) \\
\omega & \leftarrow - \frac{\gamma}{\log(1-p)} \\
\theta_{jk} & \leftarrow \frac{1}{1 + \exp\left(\beta(k-\omega)\right)} \\
d_{jk} & \sim \text{Bernoulli}(\theta_{jk})
\end{aligned}
\end{equation}\]
Simulator
Code
def prior():return dict (= np.random.exponential(scale= 1 ),= np.random.exponential(scale= 1 )def likelihood(gamma, beta, n_trials= 90 , p= 0.15 ):= - gamma / np.log1p(- p)= np.zeros(n_trials)for i in range (n_trials):= 0 while True : = 1 / (1 + np.exp(beta * (n_pumps + 1 - omega)))= np.random.binomial(n= 1 , p= theta)if not pump:break += 1 = np.random.binomial(n= 1 , p= p)if bust:break = n_pumpsreturn dict (d = d)= bf.make_simulator([prior, likelihood])
Approximator
We will use the BasicWorkflow to train an approximator that consists of an affine coupling flow inference network and a deep set summary network. The task is to infer the posterior distrobutions of \(\gamma\) and \(\delta\) , given the observed decisions to pump the baloon (\(d\) ) in each of the 90 trials.
Code
= (bf.Adapter()"d" )"gamma" , "beta" ], lower= 0 )"gamma" , "beta" ])"gamma" , "beta" ], into= "inference_variables" )"d" , "summary_variables" )
Code
= bf.BasicWorkflow(= simulator,= adapter,= bf.networks.CouplingFlow(),= bf.networks.DeepSet(),= 1e-3
Training
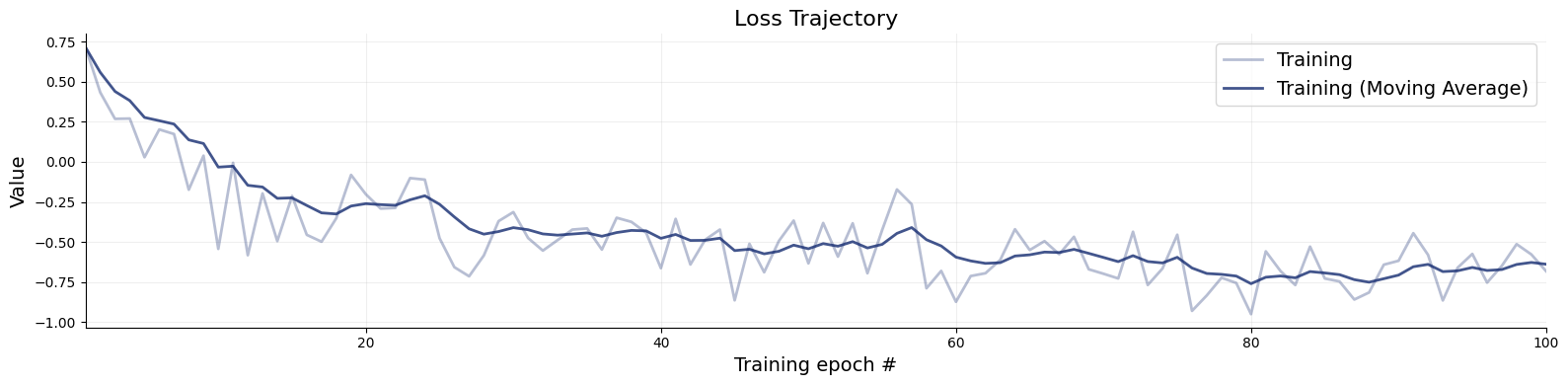
Code
= workflow.fit_online(epochs= 100 , num_batches_per_epoch= 200 , batch_size= 128 )
Validation
Code
= simulator.sample(1000 )
Code
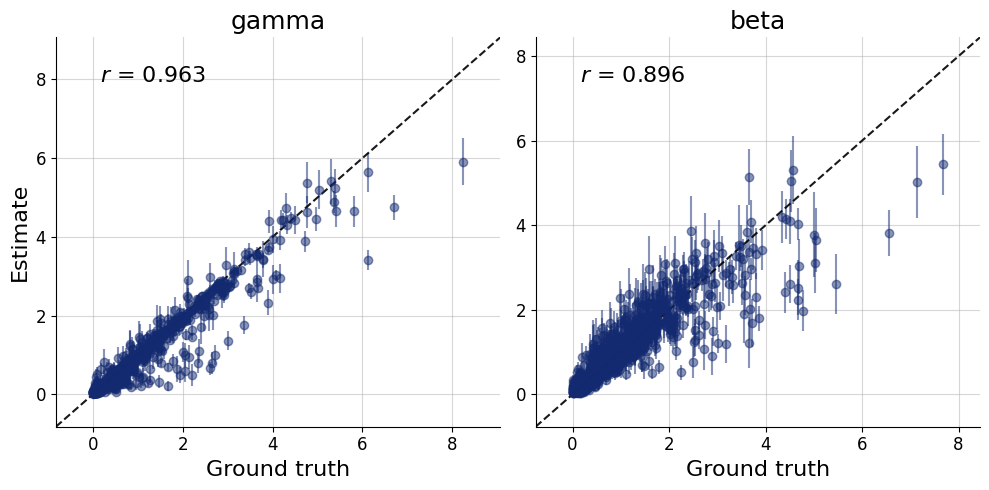
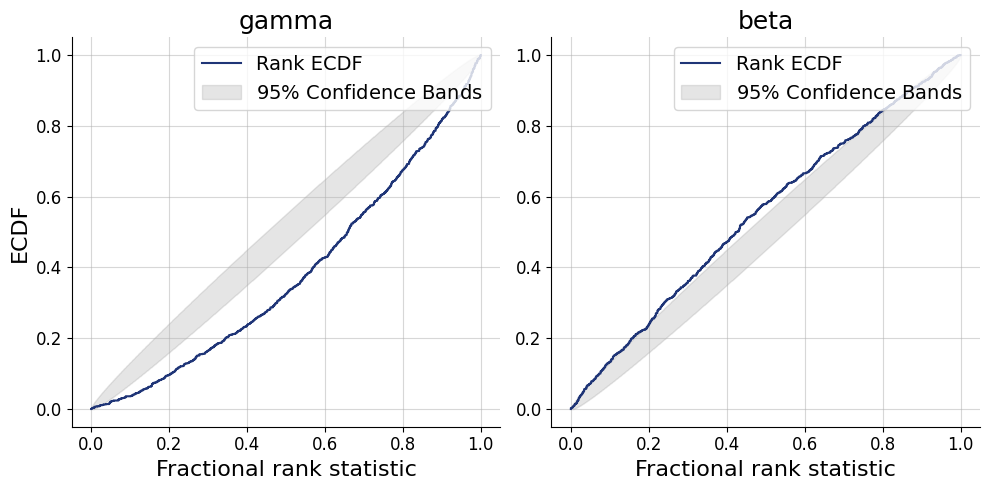
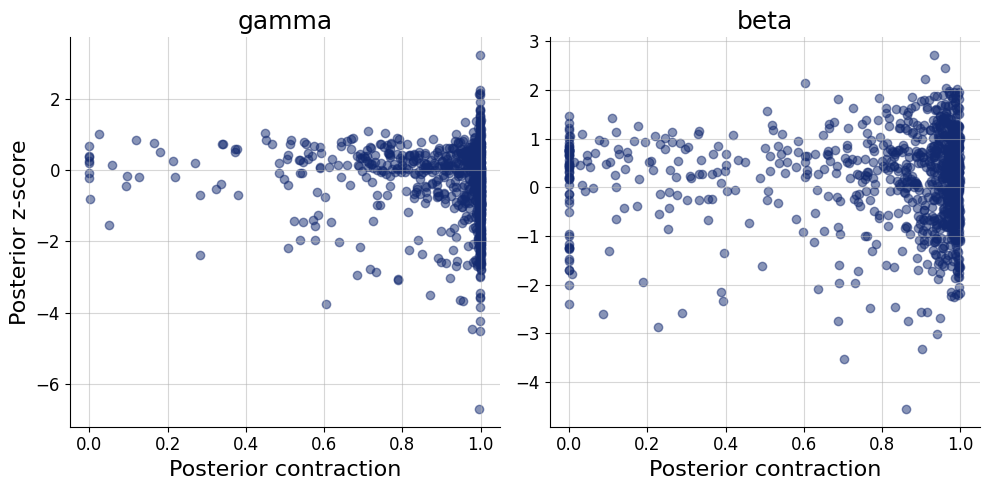
= workflow.plot_default_diagnostics(test_data= test_data, num_samples= 500 )
Inference
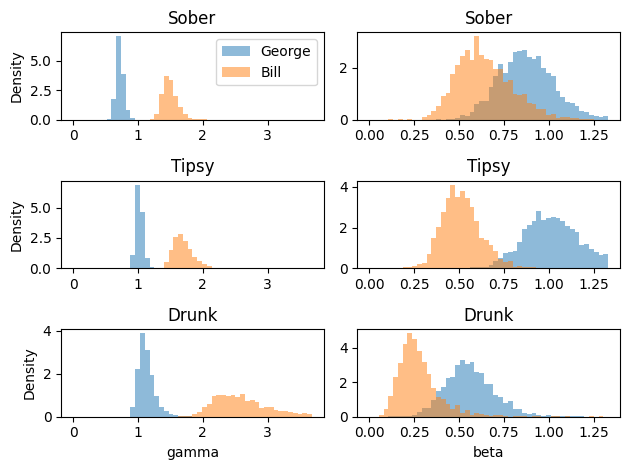
Here we will fit the model to two participants (George and Bill), each under three different conditions (Sober, Tipsy, Drunk) (Lee & Wagenmakers, 2013 ) .
Code
= np.loadtxt(os.path.join("data" , "george-bill.txt" ), delimiter= " " )= inference_data.reshape((6 , 90 , 1 ))= dict (d= inference_data)
Code
= workflow.sample(num_samples= 2000 , conditions= inference_data)
Code
= plt.subplots(3 , 2 )= {par: np.linspace(0 , np.quantile(samples, 0.99 ), 51 ) for par, samples in posterior_samples.items()}for i, cond in enumerate (["Sober" , "Tipsy" , "Drunk" ]):for j, (par, samples) in enumerate (posterior_samples.items()):= 0.5 , bins= bins[par], density= True ) + 3 ].flatten(), alpha= 0.5 , bins= bins[par], density= True ) if j == 0 :"Density" )if i == 0 :"George" , "Bill" ])if cond == "Drunk" :
References
Lee, M. D., & Wagenmakers, E.-J. (2013). Bayesian Cognitive Modeling : A Practical Course . Cambridge University Press.